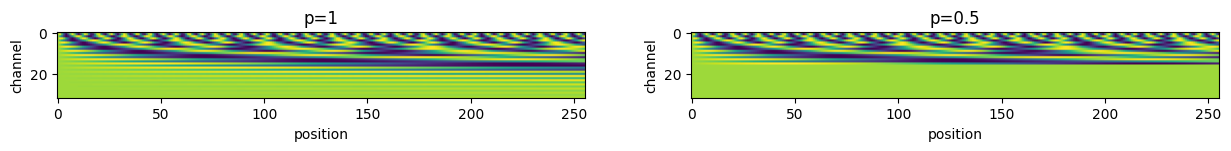
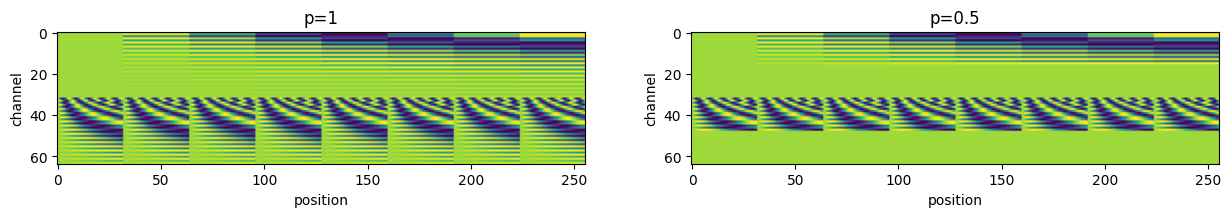
RotaryPositionalEmbedding2D
RotaryPositionalEmbedding2D (head_dim:int, p:float=1.0,
max_seq_len:int=4096, base:float=10000)
*Base class for all neural network modules.
Your models should also subclass this class.
Modules can also contain other Modules, allowing them to be nested in a tree structure. You can assign the submodules as regular attributes::
import torch.nn as nn
import torch.nn.functional as F
class Model(nn.Module):
def __init__(self) -> None:
super().__init__()
self.conv1 = nn.Conv2d(1, 20, 5)
self.conv2 = nn.Conv2d(20, 20, 5)
def forward(self, x):
x = F.relu(self.conv1(x))
return F.relu(self.conv2(x))Submodules assigned in this way will be registered, and will also have their parameters converted when you call :meth:to, etc.
.. note:: As per the example above, an __init__() call to the parent class must be made before assignment on the child.
:ivar training: Boolean represents whether this module is in training or evaluation mode. :vartype training: bool*
= 1 = 256 = 1 = 64 = torch.ones((b, s, n_heads, head_dim))= 32 = 8 = torch.arange(nx).expand(ny, - 1 )= torch.arange(ny).unsqueeze(- 1 ).expand(- 1 , nx)= torch.stack([py, px], dim=- 1 ).reshape(- 1 , 2 )= 1 = 0.5 = 100 = RotaryPositionalEmbedding2D(head_dim, p1, base= base)= pe(q, pos_idx).squeeze() # [s, head_dim] = RotaryPositionalEmbedding2D(head_dim, p2, base= base)= pe(q, pos_idx).squeeze() # [s, head_dim] = plt.subplots(1 , 2 , figsize= (15 , 5 ))f"p= { p1} " )"position" )"channel" )f"p= { p2} " )"position" )"channel" )