genQC · Generative Quantum Circuits
Generating quantum circuits with diffusion models
Generative circuit synthesis
Synthesis of discrete-continuous quantum circuits.
Pre-trained models
Easy inference of pre-trained model weights.
Open-source research
Full open source research code and model weights.
“Compile 4-qubit QFT”
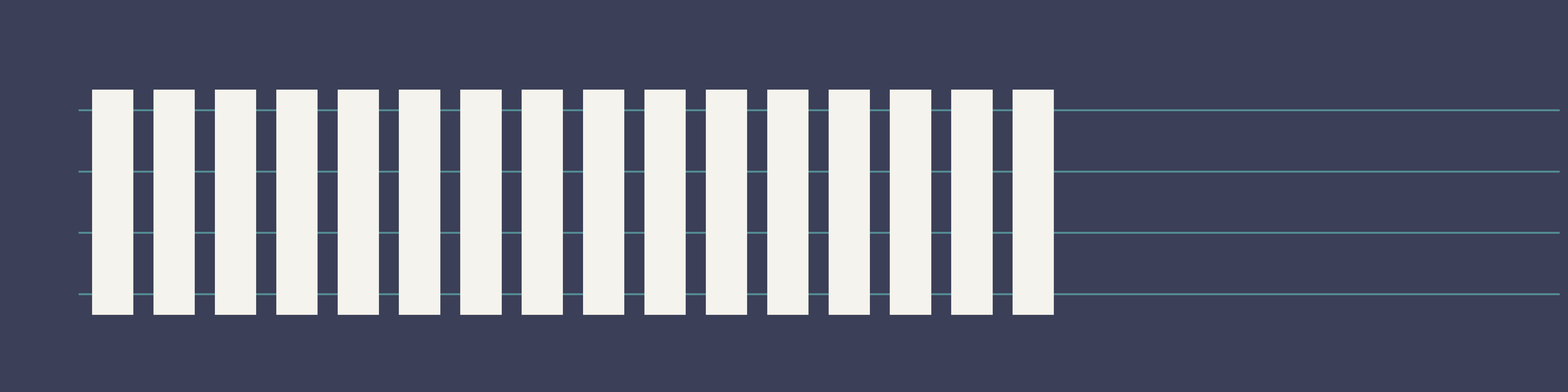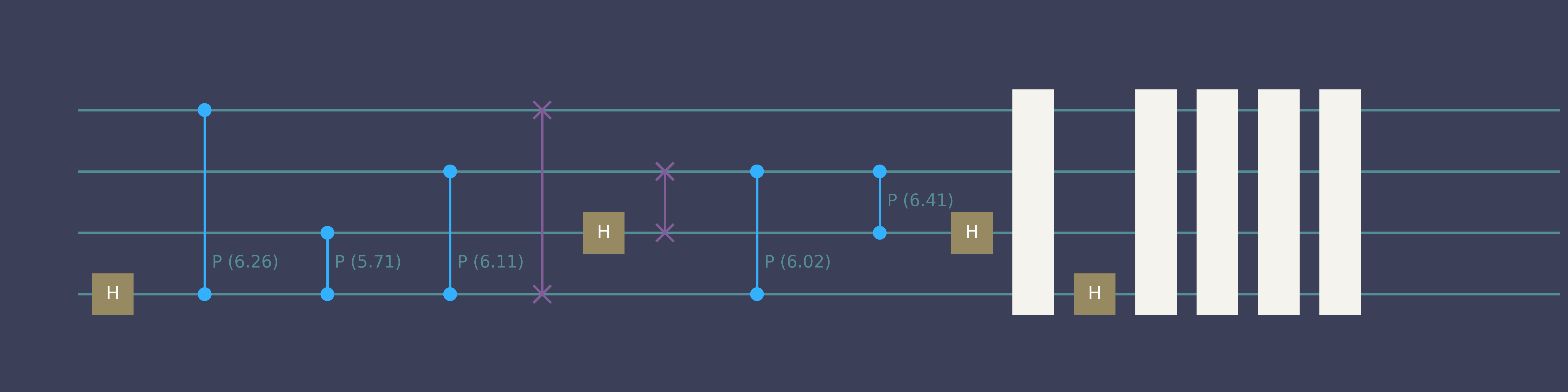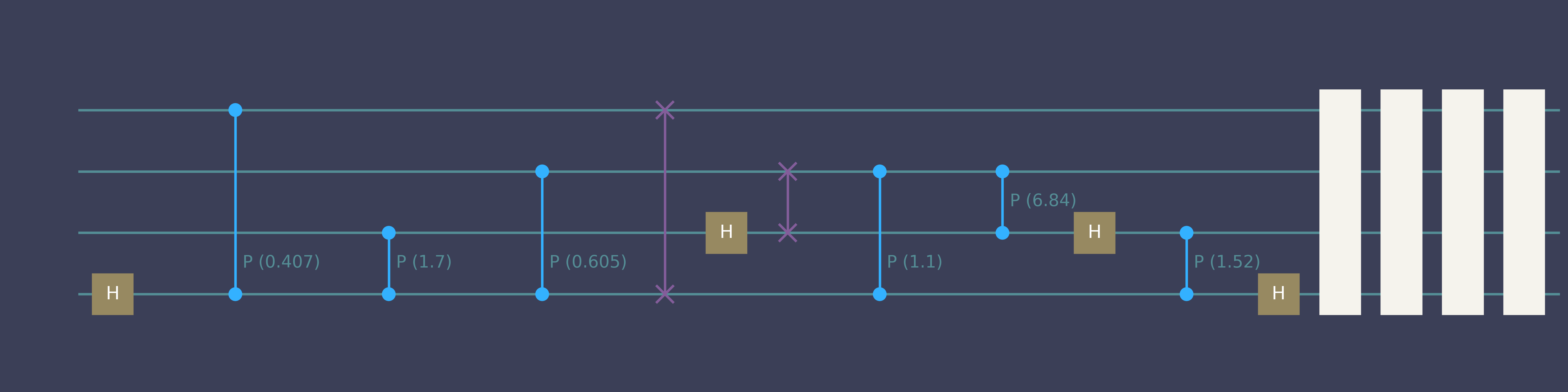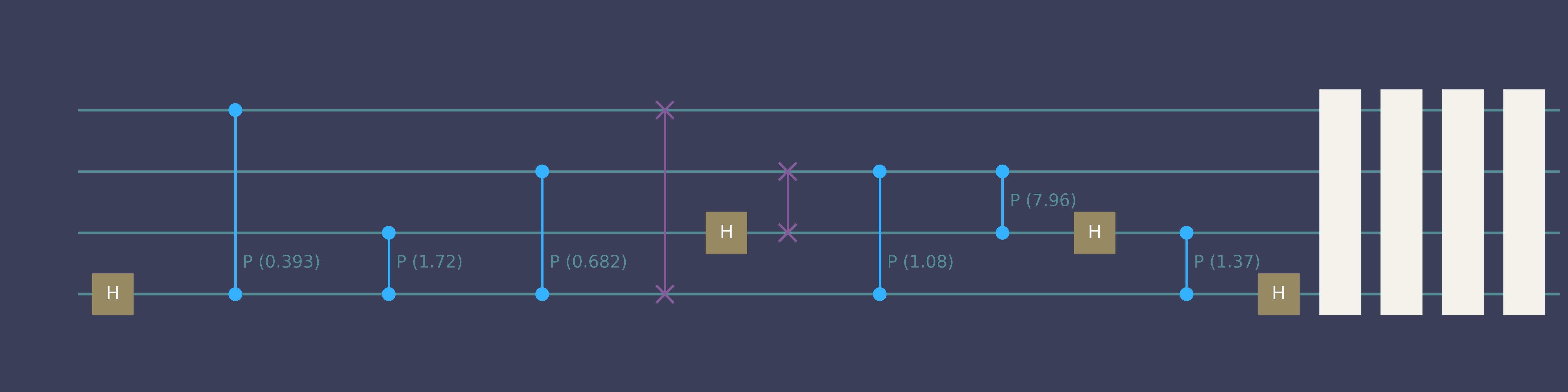
Quick start genQC
To install genQC just run:
pip install genQCOn the right hand side, a minimal example to generate a parametrized quantum circuit conditioned on the 4-qubit QFT unitary.
import torch
from genQC.pipeline.multimodal_diffusion_pipeline import MultimodalDiffusionPipeline_ParametrizedCompilation
from genQC.inference.sampling import generate_compilation_tensors, decode_tensors_to_backend
from genQC.utils.misc_utils import infer_torch_device
from genQC.platform.tokenizer.circuits_tokenizer import CircuitTokenizer
from genQC.benchmark.bench_compilation import SpecialUnitaries
from genQC.platform.simulation import Simulator, CircuitBackendType
device = infer_torch_device()
pipeline = MultimodalDiffusionPipeline_ParametrizedCompilation.from_pretrained(
repo_id="Floki00/cirdit_multimodal_compile_3to5qubit_v1.1",
device=device)
pipeline.scheduler.set_timesteps(40)
pipeline.scheduler_w.set_timesteps(40)
pipeline.g_h, pipeline.g_w = 0.3, 0.1
pipeline.lambda_h, pipeline.lambda_w = 1.0, 0.35
prompt = "Compile 4 qubits using: ['h', 'cx', 'ccx', 'swap', 'rx', 'ry', 'rz', 'cp']"
U = SpecialUnitaries.QFT(num_qubits=4).to(torch.complex64)
out_tensor, params = generate_compilation_tensors(pipeline,
prompt=prompt,
U=U,
samples=8,
system_size=5,
num_of_qubits=4,
max_gates=32)
vocabulary = {g:i+1 for i, g in enumerate(pipeline.gate_pool)}
tokenizer = CircuitTokenizer(vocabulary)
simulator = Simulator(CircuitBackendType.CUDAQ)
qc_list, _ = decode_tensors_to_backend(simulator, tokenizer, out_tensor, params)
simulator.backend.draw(qc_list[0], num_qubits=4)- 1
- Load a pre-trained Diffusion model.
- 2
- Set inference parameters.
- 3
- Specify QFT unitary.
- 4
- Generate tokenized circuits.
- 5
- Decode tensors to circuits.
- 6
- Plot a generated circuit.